Autophagy and neurodegeneration
General audience summary: Neurodegenerative diseases, such as Alzheimer’s, Parkinson’s and Huntington’s Disease, are associated with the accumulation of particular proteins that form clumps because they are not folded properly. Our research goal is to understand the links between these diseases and autophagy — the bulk recycling process that degrades proteins and particular parts of the cell. We currently focus on: understanding how autophagy is regulated using several cell and animal models; and possible ways to ramp up this process in order to remove toxic proteins and avoid the development of neurodegenerative disease.
Strategic CIMR themes: Protein Folding and Quality Control, Membrane Trafficking and Organelle Biology, Neurological Diseases, Rare Genetic Diseases
Funding: UK Dementia Research Institute programme, National Institute for Health Research - Biomedical Research Centre for Addenbrooke’s Hospital, Great Ormond Street Hospital Children's Charity, Parkinson’s UK, Tau Consortium, Merck Sharp and Dohme, Wellcome Leap, MRC
Research Group members:
Fatima Amer-Sarsour, Angeleen Fleming, Ana Lopez, Antonio Barbosa, Chun Sang, Claudia Puri, Farah Siddiqi, Jennifer Palmer, Junrui He, Lidia Wrobel, Laura Ryan, Matea Rob, Michael Takla, Pawel Obrocki, Sarah Williams, Sarayu Ramakrishnan, So Jung Park, Sungmin Son, Ye Zhu, Xinyi Li
Biography
David Rubinsztein is Professor of Molecular Neurogenetics and a UK Dementia Research Institute Group Leader at the University of Cambridge. He is Deputy Director of the Cambridge Institute for Medical Research. Dr. Rubinsztein earned his MB ChB, BSc(Med), and PhD degrees from University of Cape Town. He came to Cambridge in 1993 as a Senior Registrar in genetic pathology. His research is focused in the field of autophagy, particularly in the context of neurodegenerative diseases. His laboratory pioneered the strategy of autophagy upregulation as a possible therapeutic approach in various neurodegenerative diseases, and has identified drugs and novel pathways that may be exploited for this objective. He has made contributions that reveal the relevance of autophagy defects as a disease mechanism and to the basic cell biology of this important catabolic process. Rubinsztein was elected Fellow of the Academy of Medical Sciences (2004), EMBO member (2011), Fellow of the Royal Society (2017) and Member of Academia Europaea (2022). He was awarded the Graham Bull Prize (2007), Thudichum Medal (2017), Roger de Spoelberch prize (2017), the Goudie Medal (2020) and 2024 Movement Disorders Research Award from the American Academy of Neurology. He was identified as a Clarivates Analytics Highly Cited Researcher (2018, 2019, 2020, 2021, 2022 and 2023).
Research
Autophagy and neurodegeneration
Intracellular protein misfolding/aggregation are features of many late-onset neurodegenerative diseases, called proteinopathies. These include Alzheimer’s disease, Parkinson’s disease, tauopathies, and polyglutamine expansion diseases (like Huntington’s disease (HD) and various spinocerebellar ataxias (SCAs)). Currently, there are no effective strategies that slow/prevent the neurodegeneration resulting from these diseases in humans.
Therapeutic strategies for these diseases
The mutations causing HD and many proteinopathies confer novel toxic functions on the specific protein, and disease severity frequently correlates with expression levels. Thus, it is important to understand the factors regulating the levels of these aggregate-prone proteins.
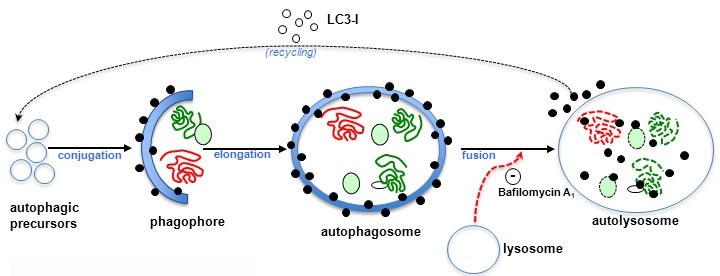
(Macro) autophagy is a bulk degradation process that mediates the clearance of long-lived proteins and organelles (reviewed in Hansen et al. (2018) Nature Reviews Molecular Cell Biology 19:579-593). Autophagosomes are formed by double-membraned structures, which engulf portions of cytoplasm. Autophagosomes ultimately fuse with lysosomes, where their contents are degraded.
Schematic overview of autophagy
We have become increasingly involved in studying autophagy, since the time of our discovery that it regulates the levels of intracytoplasmic aggregate-prone proteins that cause many neurodegenerative diseases, including Huntington’s disease, point mutations in alpha-synuclein (causing forms of Parkinson’s disease), and wild-type and mutant forms of tau (causing various dementias) (reviewed in Fleming et al. (2022) Neuron 110:935-966). The clearance of such substrates is retarded in cell models when autophagy is compromised. For Huntington’s disease and a range of related neurodegenerative diseases, we are pursuing our findings that the toxicity of these proteins in cells, Drosophila, zebrafish and mice can be alleviated by enhancing their removal by autophagy (e.g. Ravikumar et al. (2004) Nature Genetics 36: 585-595; Lopez et al. (2017) Brain 140: 1128-1146). We are trying to identify safe and effective strategies for exploiting autophagy upregulation in order to enhance the removal of the toxic intracytoplasmic proteins that cause many of these diseases (e.g. Sarkar et al. (2007) Nature Chemical Biology 3: 331-338; Williams et al. (2008) Nature Chemical Biology 4: 295-305; Vicinanza et al (2015) Molecular Cell 57:219-234; Siddiqi et al. (2019) Nature Communications 10:1817; Festa et al (2023) Neuron 111:2021-2037).
Understanding the roles of mutations
On the other hand, we have found that autophagy is compromised by mutations causing neurodegenerative diseases, including Huntington’s disease and Parkinson’s disease (recent studies: Zavodszky et al. (2014) Nature Communications 5:3828; Moreau et al. (2014) Nature Communications 5:4998; Bento et al. (2016) Nature Communications 7:11803; Pavel et al. (2016) Nature Communications 7:13821; Ashkenazi et al. (2017) Nature 545:108-111). This likely contributes to the neurotoxicity and aggregate formation seen in these conditions. Our recent data suggest that neurodegenerative disease-causing mutations in the autophagy protein WIPI4 may act to enhance cell death via ferroptosis independently of any effects on autophagy (Zhu et al (2024) Nature Cell Biology 26:542-551).
The cell biology of autophagy and its relevance to human physiology and disease, with a focus on the nervous system
Our abilities to contribute to the understanding of autophagy and neurodegenerative diseases is enhanced by our efforts to understand the signals that regulate autophagosome biogenesis (e.g. Pavel et al (2018) Nature Communications 9:2961; Nehammer, Ejlerskov, et al. (2019) eLife 2019;8:e49930; Son et al. (2019) Cell Metabolism 29:192-201; Son et al. (2020) Nature Communications 11(1):3148; Son et al (2024) Nature Cell Biology 26:235-249) and how autophagosomes are formed. Our data suggest that endocytic processes are crucial for autophagosome biogenesis (Ravikumar et al. (2010) Nature Cell Biology 12:747-757; Moreau et al. (2011) Cell 146:303-317; Puri et al. (2013) Cell 154: 1285-1299) and that autophagosomes evolve from finger-like precursor structures emerging from the RAB11A recycling endosome compartment that capture their substrates (Puri, Vicinanza et al. (2018) Developmental Cell 45: 114-131; Puri et al. (2020) Developmental Cell 53:154-168; Puri et al. (2023) Developmental Cell 58(23):2746-2760).
Publications
A Lopez, FH Siddiqi, J Villeneuve, RP Ureshino, H-Y Jeon, P Koulousakis, S Keeling , WA McEwan, A Fleming and DC Rubinsztein (2025). Carbonic anhydrase inhibition ameliorates tau toxicity via enhanced tau secretion. Nature Chemical Biology 21:577-587
SJ Park, SM Son, AD Barbosa, L Wrobel, E Stamatakou, F Squitieri, G Balmus and DC Rubinsztein (2024). Nuclear proteasomes buffer cytoplasmic proteins during autophagy compromise. Nature Cell Biology 26:1691-1699.
Y Zhu, M Fujimaki, L Snape, A Lopez, A Fleming, and DC Rubinsztein (2024). Loss of WIPI4 in neurodegeneration causes autophagy-independent ferroptosis. Nature Cell Biology 26:542-551
SM Son, SJ Park, SY Breusegem, D Larrieu and DC Rubinsztein (2024) p300 nucleocytoplasmic shuttling underlies mTORC1 hyperactivation in Hutchinson-Gilford Progeria Syndrome. Nature Cell Biology 26:235-249.
C Puri, MJ Gratian and DC Rubinsztein (2023). Mammalian autophagosomes form from finger-like phagophores. Developmental Cell 58: 2746-60
BP Festa, F H Siddiqi, M Jimenez-Sanchez, H Won, M Rob, A Djajadikerta, E Stamatakou and DC Rubinsztein (2023). Microglial-to-neuronal CCR5 signalling regulates autophagy in neurodegeneration. Neuron 111:2021-2037.
C Karibiyik, M Vicinanza, SM Son and DC Rubinsztein (2021). Glucose starvation induces autophagy via ULK1-mediated activation of PIKfyve in an AMPK-dependent manner. Developmental Cell 56:1961-1975.
SM Hill, L Wrobel, A Ashkenazi, M Fernandez-Estevez, K Tan, RW Bürli, and DC Rubinsztein (2021). VCP/p97 regulates Beclin-1-dependent autophagy initiation. Nature Chemical Biology 17:448–455
C Puri, MM Manni, M Vicinanza, C Hilcenko, Y Zhu, G Runwal, E Stamatakou, FM Menzies, K Mamchaoui, M Bitoun and DC Rubinsztein (2020). A DNM2 centronuclear myopathy mutation reveals a link between recycling endosome scission and autophagy. Developmental Cell 53:154-168
FH Siddiqi, FM. Menzies, A Lopez, E Stamatakou, C Karabiyik, R Ureshino, T Ricketts, M Jimenez-Sanchez, MA Esteban, L Lai, MD Tortorella, Z Luo, H Liu, E Metzakopian, HJR Fernandes, A Bassett, E Karran, BL Miller, A Fleming and DC Rubinsztein (2019) Felodipine induces autophagy in mouse brains with pharmacokinetics amenable to repurposing. Nature Communications 10:1817
SM Son, SJ Park, H Lee, F Siddiqi, JE Lee, FM Menzies & DC. Rubinsztein (2019) Leucine signals to mTORC1 via its metabolite acetyl-coenzyme A. Cell Metabolism 29:192-201
C Puri, M Vicinanza, A Ashkenazi, MJ Gratian, Q Zhang, CF Bento, M Renna, FM Menzies and DC Rubinsztein (2018) The RAB11A-positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P-RAB11A. Developmental Cell 45: 114-131.
A Ashkenazi, CF Bento, T Ricketts, M Vicinanza, F Siddiqi, M Pavel, F Squitieri, MC Hardenberg, S Imarisio, FM Menzies & DC Rubinsztein (2017) Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545:108-111
M Jimenez-Sanchez, et al..., and DC Rubinsztein (2015). siRNA screen identifies QPCT as a druggable target for Huntington’s disease. Nature Chemical Biology 11:347-354
M Vicinanza, VI Korolchuk, A Ashkenazi, C Puri, FM Menzies, JH Clarke, DC Rubinsztein (2015) PI(5)P regulates autophagosome biogenesis. Molecular Cell 57:219-234.
C Puri, M Renna, C Figueira Bento, K Moreau and DC Rubinsztein (2013) Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154: 1285-1299
S Luo, M Garcia-Arencibia, R Zhao, C Puri, PPC Toh, O Sadiq and DC Rubinsztein (2012) Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Molecular Cell 47:359-370
K Moreau, B Ravikumar, M Renna, C Puri, and DC Rubinsztein (2011) Autophagosome precursor maturation requires homotypic fusion. Cell 146:303-317
S Sarkar, VI Korolchuk, M Renna, S Imarisio, A Fleming, A Williams, M Garcia-Arencibia, C Rose, S Luo, BR Underwood, G Kroemer, CJ O’Kane, and DC Rubinsztein (2011) Complex inhibitory effects of nitric oxide on autophagy. Molecular Cell 43:19-32
VI Korolchuk, S Saiki, M Lichtenberg, FH Siddiqi, EA Roberts, S Imarisio, L Jahreiss, S Sarkar, M Futter, FM Menzies, CJ O’Kane, V Deretic and DC Rubinsztein (2011) Lysosomal positioning coordinates cellular nutrient responses. Nature Cell Biology 13:453-460
B Ravikumar, K Moreau, L Jahreiss, C Puri and DC Rubinsztein (2010) Plasma membrane contributes to the formation of pre-autophagosomal structures. Nature Cell Biology 12:747-757
VI Korolchuk, A Mansilla, FM Menzies and DC Rubinsztein (2009). Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Molecular Cell 33:517-527
A Williams, S Sarkar, P Cuddon, EK Ttofi, S Saiki, FH. Siddiqi, L Jahreiss, A Fleming, D Pask, P Goldsmith, CJ O’Kane, RA Floto and DC Rubinsztein (2008) Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nature Chemical Biology 4: 295-305
S Sarkar, EO Perlstein, S Imarisio, S Pineau, A Cordenier, RL Maglathlin, JA Webster, TA Lewis, CJ O’Kane, SL Schreiber*, and DC Rubinsztein* (*joint senior authors) (2007) Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nature Chemical Biology 3, 331-338.
B Ravikumar, A Acevedo-Arozena, S Imarisio, Z Berger, C Vacher, CJ O’Kane, SDM Brown and DC Rubinsztein (2005). Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nature Genetics 37:771-776.
B Ravikumar, C Vacher, Z Berger, JE Davies, S Luo, LG Oroz, F Scaravilli, DF Easton, R Duden, CJ O’Kane, and DC Rubinsztein. (2004) mTOR inhibition induces autophagy and reduces toxicity of the Huntington’s disease mutation in Drosophila and mouse models. Nature Genetics 36: 585-595
B Ravikumar, R Duden and DC Rubinsztein (2002) Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 11:1107-1117
Recent reviews:
JE Palmer, N Wilson, SM Son, P Obrocki, L Wrobel, M Rob, M Takla, VI Korolchuk, DC Rubinsztein (2025) Autophagy, ageing, and age-related neurodegeneration. Neuron 113:29-48
RA Nixon and DC Rubinsztein (2024) Mechanisms of Autophagy-Lysosome Dysfunction in Neurodegenerative Diseases. Nature Reviews Molecular Cell Biology 25:926-946.
A Fleming, M Bourdenx, M Fujimaki, C Karabiyik, GJ Krause, A Lopez, A Martín-Segura, C Puri, A Scrivo, J Skidmore, SM Son, E Stamatakou, L Wrobel, Y Zhu, AM Cuervo and DC Rubinsztein (2022). The Different Autophagy Degradation Pathways and Neurodegeneration. Neuron 110:935-966.
GM Mallucci, D Klenerman and DC Rubinsztein (2020). Developing Therapies for Neurodegenerative Disorders: Insights from Protein Aggregation and Cellular Stress Responses. Annual Review of Cell and Developmental Biology 36:165-189
M Hansen*, DC Rubinsztein*, and DW Walker* (Joint first and corresponding authors) (2018). Autophagy as a promoter of longevity – insights from model organisms. Nature Reviews Molecular Cell Biology 19:579-593