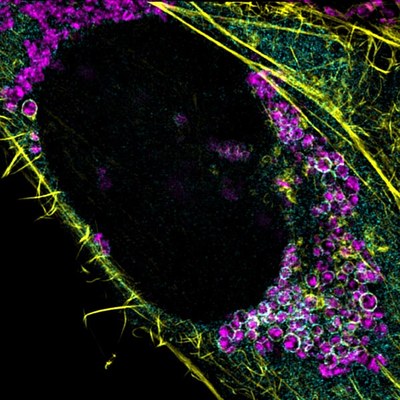
O'Loughlin T, Kruppa AJ, Ribeiro ALR, Edgar JR, Ghannam A, Smith AM, Buss F. OPTN recruitment to a Golgi-proximal compartment regulates immune signalling and cytokine secretion. J Cell Sci. 2020 Jun 15;133(12):jcs239822. doi: 10.1242/jcs.239822. PMID: 32376785; PMCID: PMC7328155.
Zakrzewski P, Lenartowska M, Buss F. Diverse functions of myosin VI in spermiogenesis. Histochem Cell Biol. 2021 Jan 2. doi: 10.1007/s00418-020-01954-x. Epub ahead of print. PMID: 33386429.
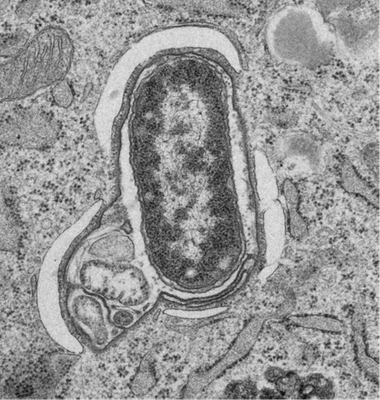
Kishi-ItakuraC, Nicholas T. Ktistakis NT, Buss F. Ultrastructural insights into pathogen clearance by autophagy. Traffic. doi: 10.1111/tra.12723 (2020).
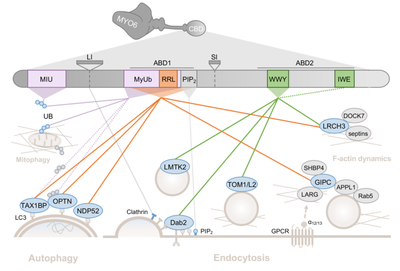
De Jonge JJ, Batters C, O’Loughlin T, Arden SD, Buss F. The MYO6 interactome: selective motor-cargo complexes fro diverse cellular processes. FEBS Lett. 593(13): 1494-1507 (2019).
O’Loughlin, T, Masters, TA and Buss, F. The MYO6 interactome reveals adaptor complexes coordinating early endosome and cytoskeletal dynamics. EMBO Reports 19(4) doi: 10.15252/embr.201744884 (2018).
Kruppa AJ, Kishi-Itakura C, Masters TA, Rorbach JE, Grice GL, Kendrick-Jones J, Nathan JA, Minczuk M and Buss F. Myosin VI-dependent actin cages encapsulate Parkin-positive damaged mitochondria. Developmental Cell 44:489-499 (2018).
Masters TA, Tumbarello DA, Chibalina MV, Buss F. MYO6 Regulates Spatial Organization of Signaling Endosomes Driving AKT Activation and Actin Dynamics. Cell Rep. 19, 2088-2101 (2017).
Brooks AB, Humphreys D, Singh V, Davidson AC, Arden SD, Buss F, Koronakis V. MYO6 is targeted by Salmonella virulence effectors to trigger PI3-kinase signaling and pathogen invasion into host cells. Proc. Natl Acad. Sci. USA 114, 3915-3920 (2017).
Masters TA, Buss F. Filopodia formation and endosome clustering induced by mutant plus-end-directed myosin VI. Proc. Natl Acad. Sci. USA 114, 1595-1600 (2017).
Tumbarello DA, Manna P, Allen M, Bycroft M, Arden SD, Kendrick-Jones J & Buss F. The autophagy receptor TAX1BP1 and the molecular motor myosin VI are required for clearance of Salmonella typhimurium by autophagy. PLoS Pathogens 11, e1005174. doi: 10.1371/journal.ppat.1005174 (2015).
Brandstaetter, H, Kishi-Itakura, C, Manstein D, Tumbarello D. and Buss F. Loss of functional myosin 1c, a motor protein involved in lipid raft trafficking, disrupts autophagosome-lysosome fusion. Autophagy 10, 2310-23 (2014).
Tumbarello, D. A., Waxse, B. J., Arden, S. D., Bright, N. A., Kendrick-Jones, J. and Buss, F. Autophagy-receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nature Cell Biol. 10, 1024–1035 (2012).